Tutorial¶
Load and visualize an SBML file.
import SBMLDiagrams
import tellurium as te
r = te.loada ('''
A -> B; v;
B -> C; v;
C -> D; v;
v = 0
''')
df = SBMLDiagrams.load(r.getSBML())
df.autolayout()
df.draw()
Load, read, edit and export to an SBML file.
import SBMLDiagrams
# The load method can access an XML string or a file name to an SBML model
df = SBMLDiagrams.load("SBML_file.xml")
df.setReactionLineThickness("reaction_id", 3.)
updated_sbmlStr = df.export()
with open('updatedModel.xml', 'w') as f:
f.write(updated_sbmlStr)
Interface to Tellurium example 1 with some basic functions, including different node shapes, dashed reaction lines, and etc.
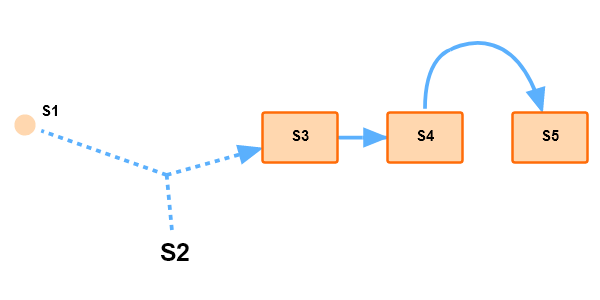
import SBMLDiagrams as sd
import tellurium as te
r = te.loada ('''
J1: S1 -> S2 + S3; k1*S1;
J2: S3 -> S4; k2*S3;
J3: S4 -> S5; k3*S4;
S1 = 10; S2 = 0;
S3 = 0; S4 = 0;
k1 = 0.1; k2 = 0.2; k3 = 0.45
''')
la = sd.load (r.getSBML())
# Arrange the network manually
la.setNodeAndTextPosition('S1', [200, 200])
la.setNodeAndTextPosition('S2', [300, 300])
la.setNodeAndTextPosition('S3', [400, 200])
la.setNodeAndTextPosition('S4', [500, 200])
la.setNodeAndTextPosition('S5', [600, 200])
# Move the node text only
la.setNodeTextPosition('S1', [200, 180])
la.setNodeTextPosition('S2', [300, 294])
la.setNodeShape('S1', 'ellipse')
la.setNodeSize('S1', [20, 20])
la.setNodeBorderWidth ('S1', 3)
la.setNodeBorderColor('S1', 'white')
la.setNodeShape('S2', 'text_only')
la.setNodeTextFontSize('S2', 20)
# Set up defaults for reaction center and bezier handles
la.setReactionDefaultCenterAndHandlePositions('J1')
la.setReactionDefaultCenterAndHandlePositions('J2')
la.setReactionDefaultCenterAndHandlePositions('J3')
la.setReactionDashStyle("J1", [5,5])
la.setReactionCenterPosition("J3",[550,150])
la.setReactionBezierHandles("J3", [[550,150],[530,155],[600,120]])
la.draw(setImageSize=[600,300])
Interface to Tellurium example 2 with alias nodes. You can assign a feature repeatly with a function.
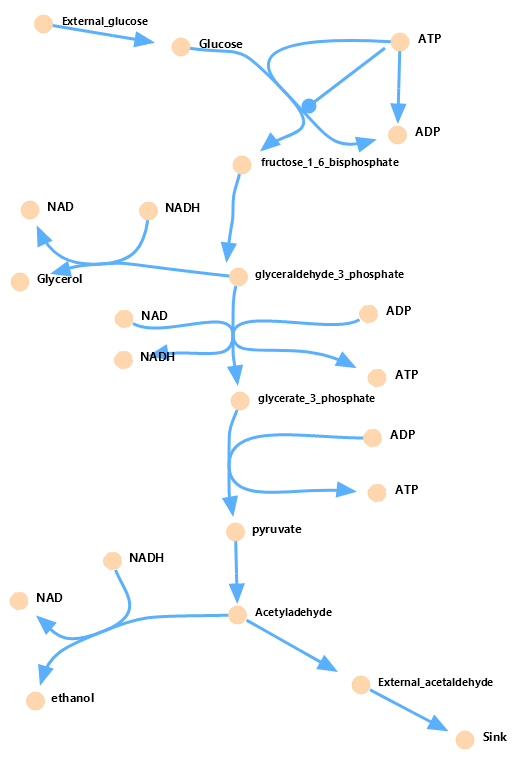
import SBMLDiagrams
import os
dirname = ""
filename = "Jana_WolfGlycolysis.xml"
with open(os.path.join(dirname, filename), 'r', encoding="utf8") as f:
sbmlStr = f.read()
la = SBMLDiagrams.load(sbmlStr)
def createCircleNode (la, id):
#get center and size of the node
num_alias = la.getNodeAliasNum(id)
if num_alias == 1:
# Change the node size and corectly adjust for the new position
center = la.getNodeCenter(id)
la.setNodeSize(id, [18, 18])
la.setNodePosition(id, [center.x-9, center.y-9])
# get the new position and size
p = la.getNodePosition(id)
size = la.getNodeSize(id)
# Position the text just outside the node
q = [p.x + 1.2*size.x, p.y-5]
la.setNodeTextPosition(id, q)
la.setNodeShape(id, 'ellipse')
la.setNodeBorderWidth (id, 0)
else:
for alias in range(num_alias):
# Change the node size and corectly adjust for the new position
center = la.getNodeCenter(id, alias = alias)
la.setNodeSize(id, [18, 18], alias = alias)
la.setNodePosition(id, [center.x-9, center.y-9], alias = alias)
# get the new position and size
p = la.getNodePosition(id, alias = alias)
size = la.getNodeSize(id, alias = alias)
# Position the text just outside the node
q = [p.x + 1.2*size.x, p.y-5]
la.setNodeTextPosition(id, q, alias = alias)
la.setNodeShape(id, 'ellipse', alias = alias)
la.setNodeBorderWidth (id, 0, alias = alias)
sp = la.getNodeIdList()
for s in sp:
createCircleNode(la, s)
la.draw(output_fileName = 'output.png')
Interface to SBGN example 1 with a complex species.
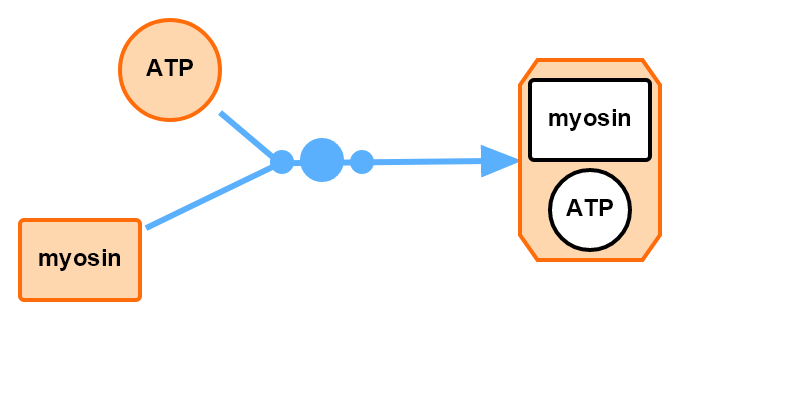
import SBMLDiagrams
import tellurium as te
r = te.loada ('''
J1: ATP + myosin -> myosinATP; k1*ATP*myosin;
ATP = 10; myosin = 10; myosinATP = 0
k1 = 0.1;
''')
sbmlStr = r.getSBML()
df = SBMLDiagrams.load(sbmlStr)
df.setNodeAndTextPosition("ATP",[100,100])
df.setNodeAndTextPosition("myosin",[50,200])
df.setNodeAndTextPosition("myosinATP",[300,120])
df.setNodeShape("ATP","ellipse")
df.setNodeAndTextSize("ATP",[50,50])
df.setNodeAndTextSize("myosinATP",[70,100])
df.setNodeTextContent("myosinATP", "-")
df.setNodeArbitraryPolygonShape("myosinATP","myosinATP-polygon", [[12.5,0],[87.5,0],[100,12.5],[100,87.5],
[87.5,100],[12.5,100],[0,87.5],[0,12.5]])
df.setReactionDefaultCenterAndHandlePositions('J1')
df.addRectangle("myosinATP_ATP", [305,130], [60,40])
df.addEllipse("myosinATP_myosin", [315,175], [40,40])
df.addText("text_myosin", "myosin", [305,130], [60,40])
df.addText("text_myosin", "ATP", [315,175], [40,40])
#print(df.getReactionCenterPosition("J1"))
#print(df.getReactionFillColor("J1"))
df.addEllipse("left_small_circle", [176.0, 166.], [10,10],
fill_color=[91, 176, 253], border_color = [91,176,253])
df.addEllipse("right_small_circle", [216.0, 166.], [10,10],
fill_color=[91, 176, 253], border_color = [91,176,253])
df.addEllipse("middle_big_circle", [191.0, 160.], [20,20],
fill_color=[91, 176, 253], border_color = [91,176,253])
df.draw(output_fileName = 'output-SBGN1.png', scale = 2)
Interface to SBGN example 2 with a gradient node.
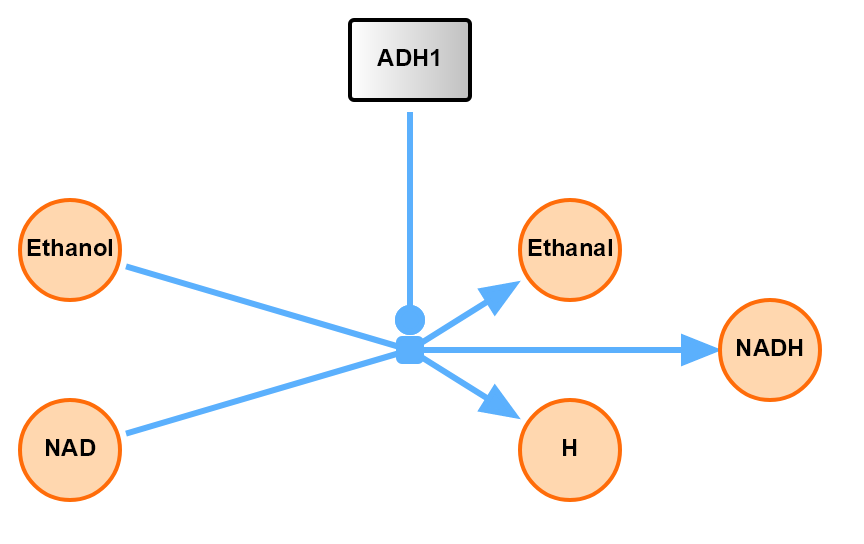
import SBMLDiagrams
import tellurium as te
r = te.loada ('''
J0: Ethanol + NAD -> Ethanal + H + NADH; k1*Ethanol*NAD/ADH1;
i1: ADH1 -| J0;
Ethanol = 10; NAD = 6; H = 0; NADH = 0; ADH1 = 5;
k1 = 0.1;
''')
sbmlStr = r.getSBML()
df = SBMLDiagrams.load(sbmlStr)
df.setNodeAndTextPosition("ADH1",[215,110])
df.setNodeAndTextPosition("Ethanol",[50,200])
df.setNodeAndTextPosition("NAD",[50,300])
df.setNodeAndTextPosition("Ethanal",[300,200])
df.setNodeAndTextPosition("H",[300,300])
df.setNodeAndTextPosition("NADH",[400,250])
df.setNodeShape("Ethanol","ellipse")
df.setNodeShape("NAD","ellipse")
df.setNodeShape("Ethanal","ellipse")
df.setNodeShape("H","ellipse")
df.setNodeShape("NADH","ellipse")
df.setNodeAndTextSize("Ethanol",[50,50])
df.setNodeAndTextSize("NAD",[50,50])
df.setNodeAndTextSize("Ethanal",[50,50])
df.setNodeAndTextSize("H",[50,50])
df.setNodeAndTextSize("NADH",[50,50])
df.setReactionDefaultCenterAndHandlePositions('J0')
#print(df.getReactionCenterPosition("J0"))
df.addRectangle("centroid_square", [239, 269], [12,12],
fill_color=[91, 176, 253], border_color = [91,176,253])
df.setNodeFillLinearGradient("ADH1", [[0.0, 50.], [100.0, 50.0]],
[[0.0, [255, 255, 255, 255]], [100.0, [192, 192, 192, 255]]])
df.setNodeBorderColor("ADH1", "black")
df.draw(output_fileName = 'output-SBGN2.png', scale = 2)
Interface to color style, i.e. loading the color style information from a JSON file.
import SBMLDiagrams
import tellurium as te
colors = SBMLDiagrams.loadColorStyle("style.json")
r = te.loada('''
A -> B; k1*A
B -> C; k2*B
k1 = 0.1; k2 = 0.2; A = 10
''')
sbmlStr = r.getSBML()
df = SBMLDiagrams.load(sbmlStr)
df.setColorStyle(colors["simplicity"])
df.draw(output_fileName="load_in_json_style/simplicity-color.png")
df.setColorStyle(colors["skyblue"])
df.draw(output_fileName="load_in_json_style/skyblue-color.png")
The file style.json:
{
"colorStyle": [
{
"style_name": "simplicity",
"compartment_fill_color": [255, 255, 255, 255],
"compartment_border_color": [255, 255, 255, 255],
"species_fill_color": [255, 255, 255, 255],
"species_border_color": [0, 0, 0, 255],
"reaction_line_color": [0, 0, 0, 255],
"lineending_fill_color": [0, 0, 0, 255],
"lineending_border_color": [0, 0, 0, 255],
"font_color": [0, 0, 0, 255],
"progress_bar_fill_color": [255, 108, 9, 200],
"progress_bar_full_fill_color": [91, 176, 253, 200],
"progress_bar_border_color": [255, 204, 153, 200]
},
{
"style_name": "skyblue",
"compartment_fill_color": [3, 219, 252, 255],
"compartment_border_color": [3, 219, 252, 255],
"species_fill_color": [23, 107, 252, 255],
"species_border_color": [119, 3, 252, 255],
"reaction_line_color": [3, 252, 157, 255],
"lineending_fill_color": [3, 252, 157, 255],
"lineending_border_color": [3, 252, 157, 255],
"font_color": [0, 0, 0, 255],
"progress_bar_fill_color": [255, 108, 9, 200],
"progress_bar_full_fill_color": [91, 176, 253, 200],
"progress_bar_border_color": [255, 204, 153, 200]
}
]
}
Interface to animation.
import SBMLDiagrams
import tellurium as te
import os
r = te.loada('''
//Created by libAntimony v2.5
model *Jana_WolfGlycolysis()
// Compartments and Species:
compartment compartment_;
species Glucose in compartment_, fructose_1_6_bisphosphate in compartment_;
species glyceraldehyde_3_phosphate in compartment_, glycerate_3_phosphate in compartment_;
species pyruvate in compartment_, Acetyladehyde in compartment_, External_acetaldehyde in compartment_;
species ATP in compartment_, ADP in compartment_, NAD in compartment_, NADH in compartment_;
species $External_glucose in compartment_, $ethanol in compartment_, $Glycerol in compartment_;
species $Sink in compartment_;
// Reactions:
J0: $External_glucose => Glucose; J0_inputFlux;
J1: Glucose + 2ATP => fructose_1_6_bisphosphate + 2ADP; J1_k1*Glucose*ATP*(1/(1 + (ATP/J1_Ki)^J1_n));
J2: fructose_1_6_bisphosphate => glyceraldehyde_3_phosphate + glyceraldehyde_3_phosphate; J2_J2_k*fructose_1_6_bisphosphate;
J3: glyceraldehyde_3_phosphate + NADH => NAD + $Glycerol; J3_J3_k*glyceraldehyde_3_phosphate*NADH;
J4: glyceraldehyde_3_phosphate + ADP + NAD => ATP + glycerate_3_phosphate + NADH; (J4_kg*J4_kp*glyceraldehyde_3_phosphate*NAD*ADP - J4_ka*J4_kk*glycerate_3_phosphate*ATP*NADH)/(J4_ka*NADH + J4_kp*ADP);
J5: glycerate_3_phosphate + ADP => ATP + pyruvate; J5_J5_k*glycerate_3_phosphate*ADP;
J6: pyruvate => Acetyladehyde; J6_J6_k*pyruvate;
J7: Acetyladehyde + NADH => NAD + $ethanol; J7_J7_k*Acetyladehyde*NADH;
J8: Acetyladehyde => External_acetaldehyde; J8_J8_k1*Acetyladehyde - J8_J8_k2*External_acetaldehyde;
J9: ATP => ADP; J9_J9_k*ATP;
J10: External_acetaldehyde => $Sink; J10_J10_k*External_acetaldehyde;
// Species initializations:
Glucose = 0;
fructose_1_6_bisphosphate = 0;
glyceraldehyde_3_phosphate = 0;
glycerate_3_phosphate = 0;
pyruvate = 0;
Acetyladehyde = 0;
External_acetaldehyde = 0;
ATP = 3;
ADP = 1;
NAD = 0.5;
NADH = 0.5;
External_glucose = 0;
ethanol = 0;
Glycerol = 0;
Sink = 0;
// Compartment initializations:
compartment_ = 1;
// Variable initializations:
J0_inputFlux = 50;
J1_k1 = 550;
J1_Ki = 1;
J1_n = 4;
J2_J2_k = 9.8;
J3_J3_k = 85.7;
J4_kg = 323.8;
J4_kp = 76411.1;
J4_ka = 57823.1;
J4_kk = 23.7;
J5_J5_k = 80;
J6_J6_k = 9.7;
J7_J7_k = 2000;
J8_J8_k1 = 375;
J8_J8_k2 = 375;
J9_J9_k = 28;
J10_J10_k = 80;
J2_k = 9.8;
J3_k = 85.7;
J5_k = 80;
J6_k = 9.7;
J7_k = 2000;
J8_k1 = 375;
J8_k2 = 375;
J9_k = 28;
J10_k = 80;
//Other declarations:
const compartment_, J0_inputFlux, J1_k1, J1_Ki, J1_n, J2_J2_k, J3_J3_k;
const J4_kg, J4_kp, J4_ka, J4_kk, J5_J5_k, J6_J6_k, J7_J7_k, J8_J8_k1, J8_J8_k2;
const J9_J9_k, J10_J10_k, J2_k, J3_k, J5_k, J6_k, J7_k, J8_k1, J8_k2, J9_k;
const J10_k;
end
''')
DIR = os.path.dirname(os.path.abspath(__file__))
filename = "Jana_WolfGlycolysis.xml"
f = open(os.path.join(DIR, filename), 'r')
sbmlStr = f.read()
f.close()
SBMLDiagrams.animate(0,30,1000, r, 0.5, sbmlStr=sbmlStr, outputName="output")